Peripheral Blood CD4+ Helper T Cells, Frozen
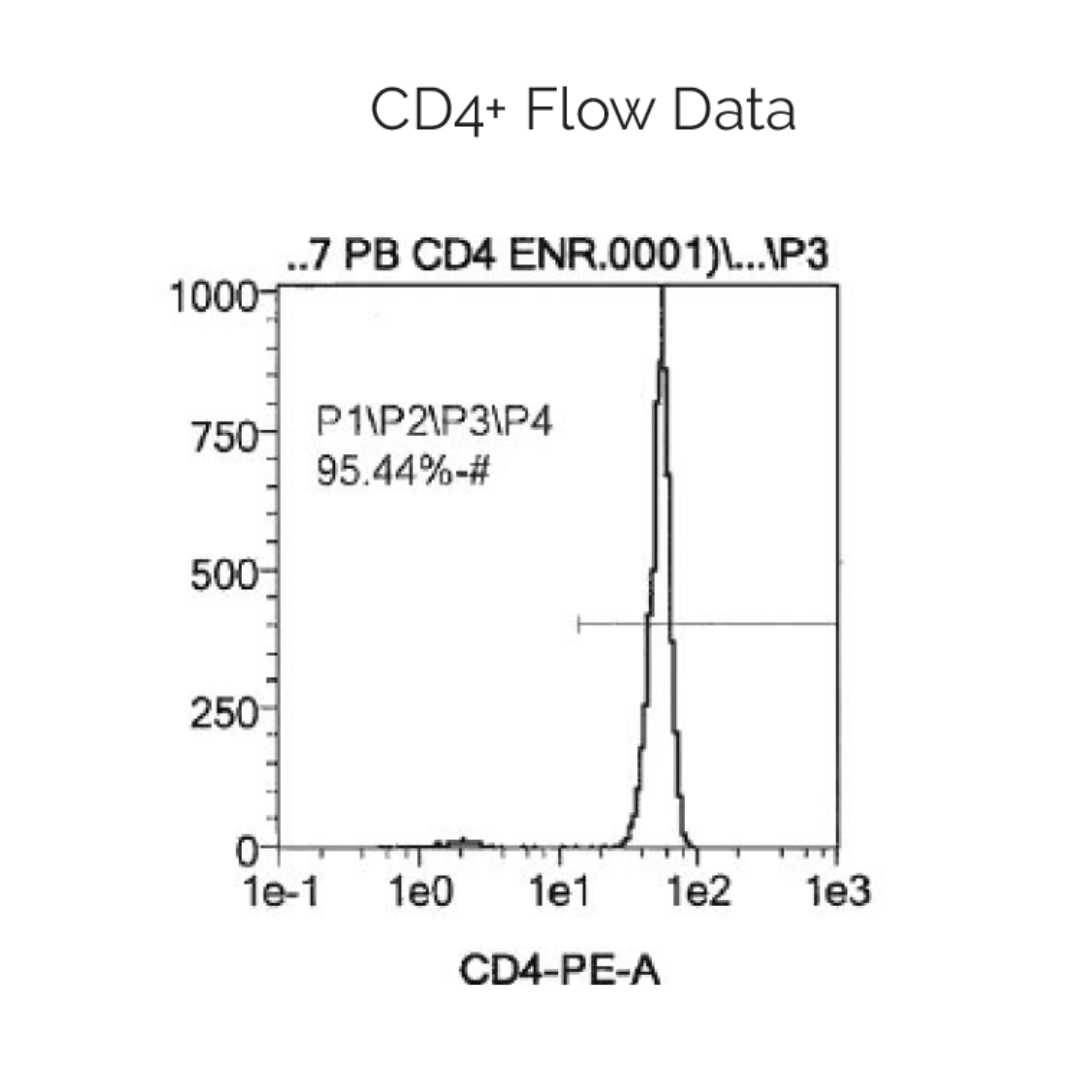
Human peripheral blood CD4+ helper T cells are enriched by means of negative selection. Cells expressing CD8, CD14, CD15, CD16, CD19, CD36, CD56, CD123, TCRγ/δ, and CD235a are depleted from the peripheral blood mononuclear cell population using immunomagnetic particles leaving purified, untouched CD4+ helper T cells. Cell isolations are characterized by flow cytometry to ensure a highly pure and viable cell population.
Cells were obtained using Institutional Review Board (IRB) approved consent forms and protocols.
Learn more about bulk ordering through CGT Global HERE.
Since our founding in 2010, CGT Global has pursued our mission to transform healthcare as we accelerate cell and gene therapy research and clinical trials, streamline the commercialization of new treatments, and map the last mile to patient access to these life-changing remedies. By innovating each stage in the cycle; development, commercialization, and delivery, we reduce the overall cost of the care and multiply access points so that millions can receive cutting edge, life-saving gene and cell therapies.

Description
Human CD4+ helper T cells express the CD4 co-receptor and play an important role in the adaptive immune system by assisting other white blood cells. These cells are activated when they are presented an antigen on the MHC class II molecule of an antigen-presenting cell (APC). Once activated, CD4+ helper T cells secrete lymphokines that assist in the activation of B cells to secrete antibodies that target specific microbes and assist in the activation of cytotoxic T cells to destroy infected target cells.
Additional information
| Anticoagulant | |
|---|---|
| Format | |
| Grade | |
| Species | |
| Cell and Tissue Source | |
| Disease State | |
| Donor Attributes |
Schillebeeckx et al. (2020) Analytical Performance of an Immunoprofiling Assay Based on RNA Models. J Mol Diagn. doi.org/10.1016/j.jmoldx.2020.01.009
Joberty et al. (2020) A Tandem Guide RNA-Based Strategy for Efficient CRISPR Gene Editing of Cell Populations with Low Heterogeneity of Edited Alleles. The CRISPR Journal. 3(2): 123-134.